-
Faculty of Arts and HumanitiesDean's Office, Faculty of Arts and HumanitiesJakobi 2, r 116-121 51005 Tartu linn, Tartu linn, Tartumaa EST0Institute of History and ArchaeologyJakobi 2 51005 Tartu linn, Tartu linn, Tartumaa EST0Institute of Estonian and General LinguisticsJakobi 2, IV korrus 51005 Tartu linn, Tartu linn, Tartumaa EST0Institute of Philosophy and SemioticsJakobi 2, III korrus, ruumid 302-337 51005 Tartu linn, Tartu linn, Tartumaa EST0Institute of Cultural ResearchÜlikooli 16 51003 Tartu linn, Tartu linn, Tartumaa EST0Institute of Foreign Languages and CulturesLossi 3 51003 Tartu linn, Tartu linn, Tartumaa EST0School of Theology and Religious StudiesÜlikooli 18 50090 Tartu linn, Tartu linn, Tartumaa EST0Viljandi Culture AcademyPosti 1 71004 Viljandi linn, Viljandimaa EST0Professors emeriti, Faculty of Arts and Humanities0Associate Professors emeriti, Faculty of Arts and Humanities0Faculty of Social SciencesDean's Office, Faculty of Social SciencesLossi 36 51003 Tartu linn, Tartu linn, Tartumaa EST0Institute of EducationJakobi 5 51005 Tartu linn, Tartu linn, Tartumaa EST0Johan Skytte Institute of Political StudiesLossi 36, ruum 301 51003 Tartu linn, Tartu linn, Tartumaa EST0School of Economics and Business AdministrationNarva mnt 18 51009 Tartu linn, Tartu linn, Tartumaa EST0Institute of PsychologyNäituse 2 50409 Tartu linn, Tartu linn, Tartumaa EST0School of LawNäituse 20 - 324 50409 Tartu linn, Tartu linn, Tartumaa EST0Institute of Social StudiesLossi 36 51003 Tartu linn, Tartu linn, Tartumaa EST0Narva CollegeRaekoja plats 2 20307 Narva linn, Ida-Virumaa EST0Pärnu CollegeRingi 35 80012 Pärnu linn, Pärnu linn, Pärnumaa EST0Professors emeriti, Faculty of Social Sciences0Associate Professors emeriti, Faculty of Social Sciences0Faculty of MedicineDean's Office, Faculty of MedicineRavila 19 50411 Tartu linn, Tartu linn, Tartumaa ESTInstitute of Biomedicine and Translational MedicineBiomeedikum, Ravila 19 50411 Tartu linn, Tartu linn, Tartumaa ESTInstitute of PharmacyNooruse 1 50411 Tartu linn, Tartu linn, Tartumaa ESTInstitute of DentistryL. Puusepa 1a 50406 Tartu linn, Tartu linn, Tartumaa ESTInstitute of Clinical MedicineL. Puusepa 8 50406 Tartu linn, Tartu linn, Tartumaa ESTInstitute of Family Medicine and Public HealthRavila 19 50411 Tartu linn, Tartu linn, Tartumaa ESTInstitute of Sport Sciences and PhysiotherapyUjula 4 51008 Tartu linn, Tartu linn, Tartumaa ESTProfessors emeriti, Faculty of Medicine0Associate Professors emeriti, Faculty of Medicine0Faculty of Science and TechnologyDean's Office, Faculty of Science and TechnologyVanemuise 46 - 208 51003 Tartu linn, Tartu linn, Tartumaa ESTInstitute of Computer ScienceNarva mnt 18 51009 Tartu linn, Tartu linn, Tartumaa ESTInstitute of GenomicsRiia 23b/2 51010 Tartu linn, Tartu linn, Tartumaa ESTEstonian Marine Institute0Institute of PhysicsInstitute of ChemistryRavila 14a 50411 Tartu linn, Tartu linn, Tartumaa EST0Institute of Mathematics and StatisticsNarva mnt 18 51009 Tartu linn, Tartu linn, Tartumaa EST0Institute of Molecular and Cell BiologyRiia 23, 23b - 134 51010 Tartu linn, Tartu linn, Tartumaa ESTTartu ObservatoryObservatooriumi 1 61602 Tõravere alevik, Nõo vald, Tartumaa EST0Institute of TechnologyNooruse 1 50411 Tartu linn, Tartu linn, Tartumaa ESTInstitute of Ecology and Earth SciencesJ. Liivi tn 2 50409 Tartu linn, Tartu linn, Tartumaa ESTProfessors emeriti, Faculty of Science and Technology0Associate Professors emeriti, Faculty of Science and Technology0Institute of BioengineeringArea of Academic SecretaryHuman Resources OfficeUppsala 6, Lossi 36 51003 Tartu linn, Tartu linn, Tartumaa EST0Area of Head of FinanceFinance Office0Area of Director of AdministrationInformation Technology Office0Administrative OfficeÜlikooli 17 (III korrus) 51005 Tartu linn, Tartu linn, Tartumaa EST0Estates Office0Marketing and Communication OfficeÜlikooli 18, ruumid 102, 104, 209, 210 50090 Tartu linn, Tartu linn, Tartumaa EST0Area of Vice Rector for DevelopmentCentre for Entrepreneurship and InnovationNarva mnt 18 51009 Tartu linn, Tartu linn, Tartumaa EST0University of Tartu Natural History Museum and Botanical GardenVanemuise 46 51003 Tartu linn, Tartu linn, Tartumaa EST0International Cooperation and Protocol Office0University of Tartu MuseumLossi 25 51003 Tartu linn, Tartu linn, Tartumaa EST0Area of RectorRector's Strategy OfficeInternal Audit OfficeArea of Vice Rector for Academic AffairsOffice of Academic AffairsUniversity of Tartu Youth AcademyUppsala 10 51003 Tartu linn, Tartu linn, Tartumaa EST0Student Union OfficeÜlikooli 18b 51005 Tartu linn, Tartu linn, Tartumaa EST0Centre for Learning and TeachingArea of Vice Rector for ResearchUniversity of Tartu LibraryW. Struve 1 50091 Tartu linn, Tartu linn, Tartumaa EST0Grant Office
Geneticists are bringing personalised medicine closer to recently admixed individuals
A new study in Nature Communications proposes a method to extend polygenic scores, the estimate of genetic risk factors and a cornerstone of the personalised medicine revolution, to individuals with multiple ancestral origins. The study was led by Dr Davide Marnetto from the Institute of Genomics of the University of Tartu, Estonia and coordinated by Dr Luca Pagani from the same institution and from the University of Padova, Italy.
“The information contained in our DNA is a mosaic of genetic instructions inherited from our ancestors, and in many societies one’s ancestors often come from the opposite corners of the world,” says Dr Davide Marnetto, the first author of the study. The contribution of one’s ancestry, or ancestries, to the total risk of developing a specific disease or presenting a given trait is a long standing question of medical genomics. Nevertheless, most of the genotype/phenotype association data come from relatively uniform populations, in order to have a simplified and clearer picture. But what can be done when dealing with individuals who derive their ancestry from two or more distantly related populations? “The latest developments of personalised medicine needed an extra step to be applied to individuals with more diverse origins, and here we tried to combine knowledge from homogeneous populations into a model that could work for recently admixed individuals,” continues Dr Marnetto.
To separate the various genomic components of each individual, Marnetto and colleagues applied methods from molecular anthropology and population genomics. “This research is a welcomed example of deep synergy between evolutionary/population genetics framework and medically oriented large scale genomics science, which is one of the focuses of our institute,” says Dr Mait Metspalu who is heading the Institute of Genomics at the University of Tartu.
“Our work provides a solid proof of principle on the feasibility of using population genetic and molecular anthropology to boost the potential of personalised medicine. I hope our work can bring individuals of mixed ancestry one step closer to the benefits of personalised and predictive healthcare,” concludes Dr Luca Pagani, the research coordinator.
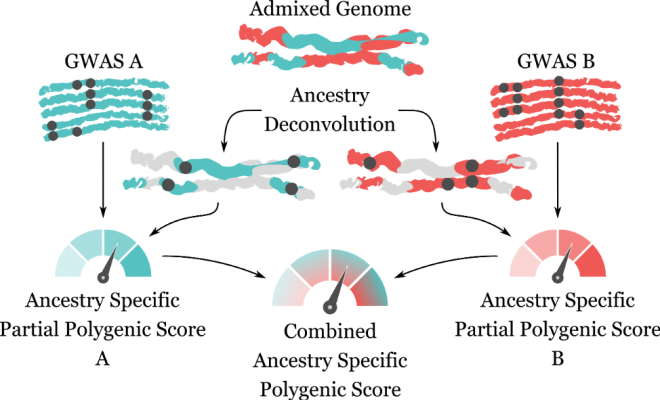
The genome from an admixed individual (blue/red) is separated into the two main ancestral components (ancestry deconvolution). Mutations associated with diseases or phenotypic traits on each component (black dots) are studied separately and two ancestry-specific partial polygenic scores (A and B) are computed using information from population-specific genome-wide association studies (GWAS). The two ancestry-specific partial polygenic scores are then combined together to obtain the individual’s ancestry-specific polygenic score. The visual was made by Davide Marnetto.
Further information:
Davide Marnetto, Research Fellow of Population Genetics at the University of Tartu Institute of Genomics, +372 5395 8881, davide.marnetto [ät] ut.ee
Luca Pagani, Senior Research Fellow of Population Genetics at the University of Tartu Institute of Genomics and Associate Professor at the University of Padova Department of Biology, +3934 0786 2518, +3904 9827 6290, luca.pagani [ät] ut.ee
Mob: +(372) 5307 7820

